Portfolio
We work with many groups both within and external to the University of Colorado:
- Greene Lab
- TISLab
- Way Lab
- JRaviLab
- Krishnan Lab
- BioThings.io
- The Wiley Lab
- …and more!
Projects
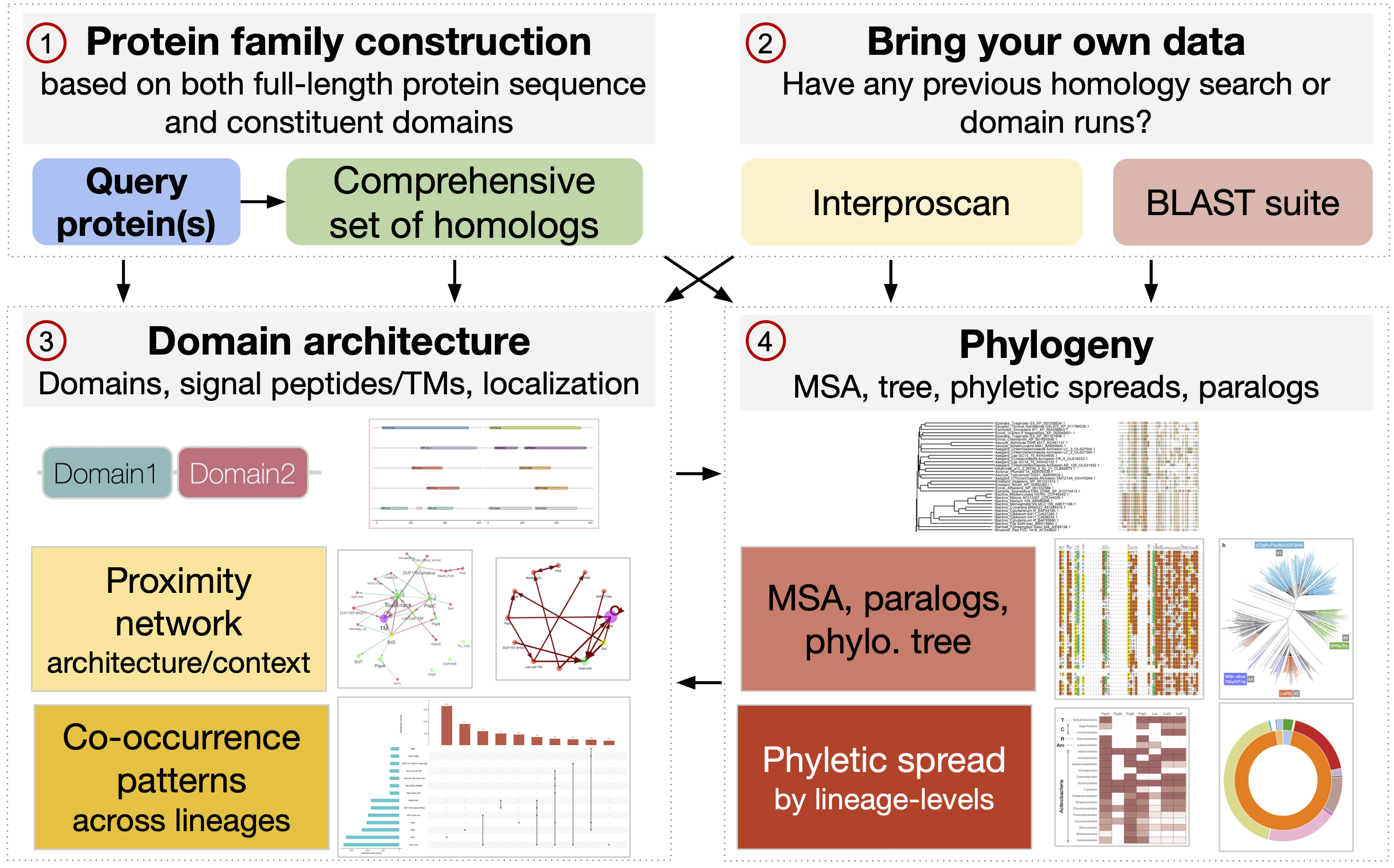
A web app that enables researchers to run a general-purpose computational workflow for characterizing the molecular evolution and phylogeny of their proteins of interest.
A suite of common functions used to process high dimensional readouts from high-throughput cell experiments.
A Python package which enables large data processing to enhance single-cell morphology data analysis.

A web app and supporting backend for simplifying scientific and medical writing.

A web app built from scratch to allow users to collect, save, and share sets of genes. A geneSET companion to MyGene.info.

A frontend web app and supporting backend server that allows users to explore how a word changes in meaning over time based on natural language processing machine learning.

A redesign and rewrite of the Monarch Initiative application from the ground up, designed to be more modern, maintainable, robust, and accessible.
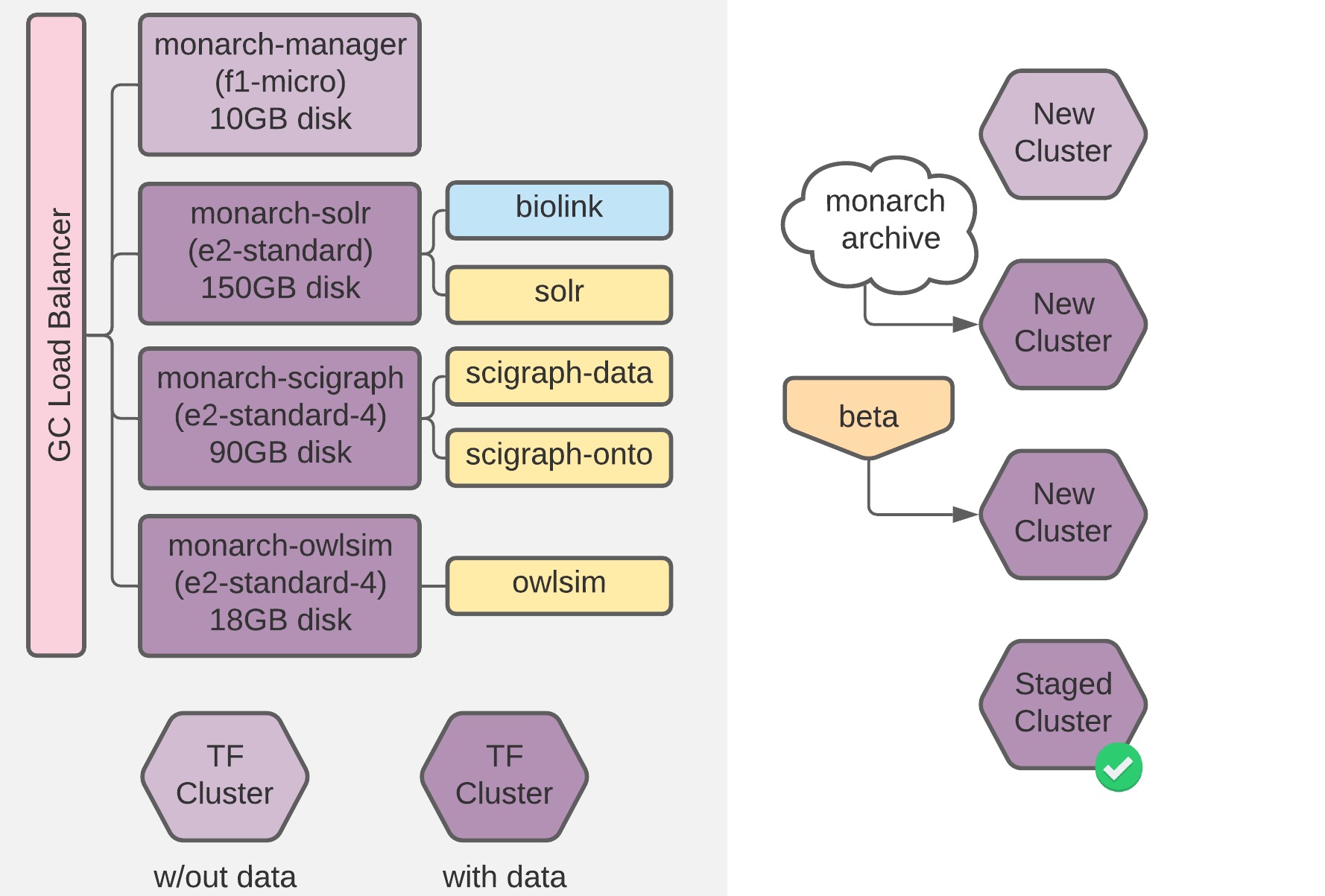
A migration of the all Monarch Initiative backend and associated services from physical hardware to Google Cloud, including automated provisioning and deployment via Terraform, Ansible, and Docker Swarm.
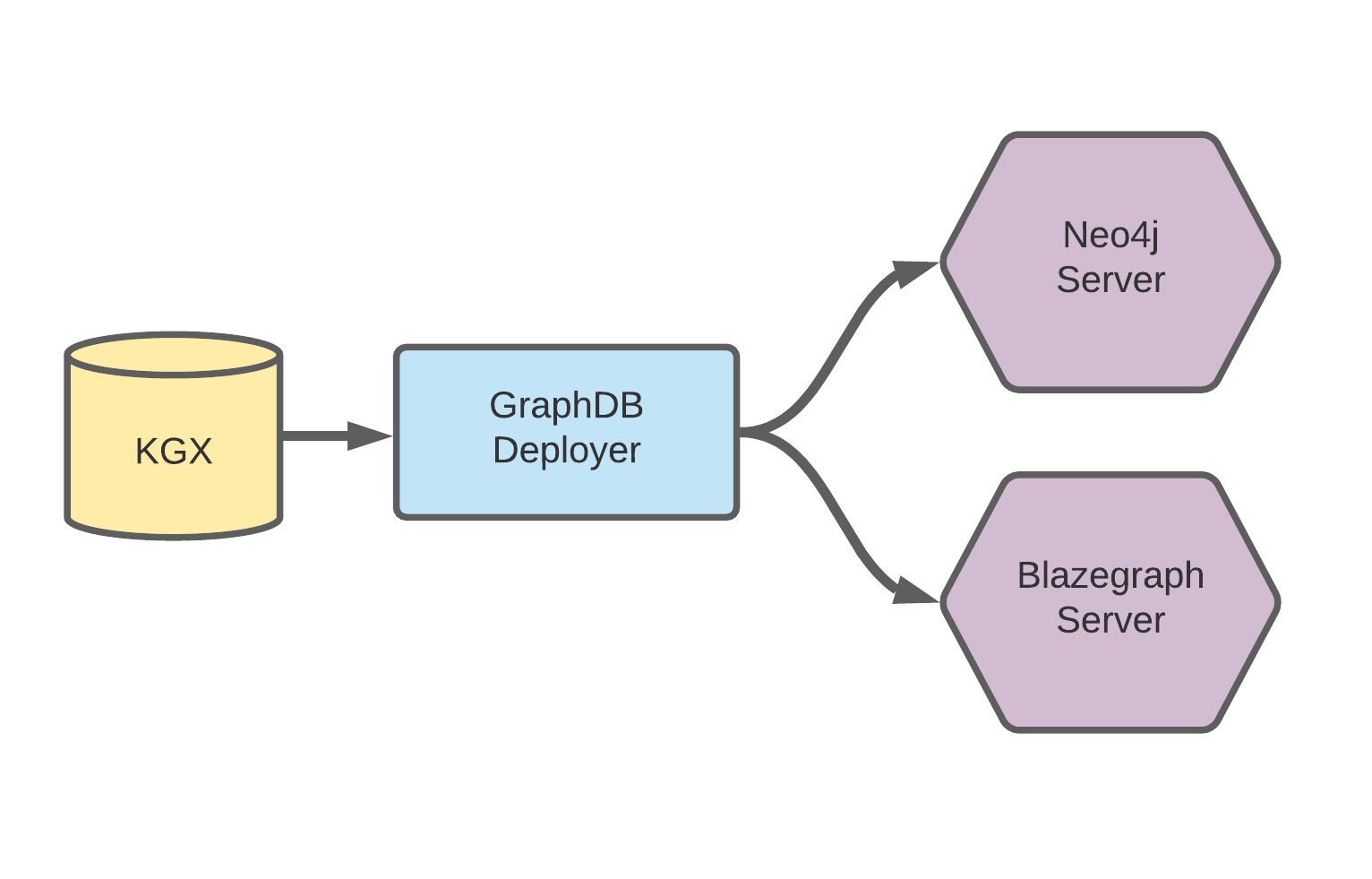
Automates the parsing and transformation of a KGX archive into graphdb-ready formats. After the archive is converted, automates provisioning and deployment of Neo4j and Blazegraph instances from a KGX archive.

An easy-to-use, flexible website template for labs. What this very site is built on!
